Patients and tissue samples
Written informed consent (provided by Springer Nature) was given by the next of kin of all patients, according to CARE guidelines and in compliance with the Declaration of Helsinki principles. It covers the permission to analyse and publish the obtained data, and explicitely states that full anonymity can not be guaranted. Clinical data have been anonymised. Sex and gender were not taken into considerations.
This study was approved by the internal review board of the University Hospital Zurich and the Cantonal Ethics Committee of Zurich, Switzerland (KEK-ZH-Nr. 2013-0504).
Patients’ clinical presentation and histopathologic findings are detailed in the Supplementary appendix.
Histopathologic evaluation of tissue samples
Formalin-fixed, paraffin-embedded (FFPE) liver and kidney specimens were processed according to standard histologic methods including hematoxylin & eosin stain (H&E), liver specimens additionally with periodic acid-Schiff after diastase digestion stain (PAS-D) as well as a connective tissue stain such as Masson trichrome, kidney specimens additionally with periodic acid-Schiff (PAS), silver methenamine stain, elastic van Gieson-stain (EVG) and acid fuchsin orange G stain (AFOG). Duodenum specimen was processed with Alcian Blue for PAS, in conjunction with PAS staining (AB/PAS) for visualization of the glycocalix.
Histologic slides including archived and newly stained slides from prior kidney biopsies and tissue gathered at autopsy were evaluated independently by experienced renal (BH, AG, HH, HY) or liver (DL, AW) pathologists, respectively.
Immunohistochemistry (IHC)
Distinct mouse monoclonal antibodies (mAbs) were used to target the different HEV ORF2 protein isoforms: 1E640 (Millipore Corporation, MAB8002, clone 1E6) and P3H222 for glycosylated (ORF2g/c) and nonglycosylated (ORF2i) isoforms, P1H1, P2H1 and P2H2 for non-glycosylated infectious HEV ORF2i only18,22. IgG isotype control antibody (Invitrogen) was used in parallel of HEV ORF2 protein staining using 1E6 antibody according to the standard procedures at the Department of Pathology and Molecular Pathology. FFPE cytospin material from Hep293TT cells replicating a full-length HEV genome HEV or the green fluorescent protein were used as positive and negative controls40. Visualization of the HEV ORF2 capsid protein was achieved using 1E6 antibody (as described in ref. 40) and P1H1, P2H1, P2H2 or P3H2 antibodies at a 1/250 dilution including a 90 min pretreatment at 100 °C with buffer CC1 and direct detection with OptiView Kit (Ventana). For further immunohistochemistry on FFPE sections, standard procedures at the Department of Pathology and Molecular Pathology were applied using the Ventana BenchMark automated staining system with IgG, IgA, IgM, and C3 antibodies (Dako, A0424, P0216, A0426 and A0062, dilution: 1:45 000, 1:30 000, 1:7 500 and 1:2 000 respectively).
On-slide deglycosylation
For the on-slide deglycosylation experiments, the protocol was adapted from Wang et al.43. The sections were deparaffinized, processed for antigen retrieval step (as required for the IHC protocol using 1E6 antibody40) and incubated with a deglycosylation mix composed of: protein deglycosylation mix II, beta 1-3 galactosidase and alpha 1-3, 4, 6 galactosidase (New England Biolabs (NEB), P6044Sm P0726S and P0747S respectively), at RT for 30 minutes followed by a 16 h incubation at 37 °C. The sections were then further stained with 1E6 antibody as described in ref. 40. As control, FFPE sections of human duodenum were stained with AB/PAS preceded or not by an on-slide deglycosylation step to assess the effect on the glycolalyx.
Immunofluorescence
For IgG/HEV ORF2 co-staining, we used either fresh frozen (6 µm thick) or FFPE (2 µm thick) tissue sections from kidney transplant tissue. Sections were mounted on glass slides (SuperFrost Gold) and dried for 30 min at 37 °C.
Fresh frozen sections were fixed in chemically pure acetone for 10 minutes. Slides were then air dried, pretreated for 5 minutes with a solution of 0.1% polyoxyethylene (20) sorbitan monolaurate (Tween 20) in tris-buffered-saline and washed with distilled H2O. FFPE tissue section were deparaffinized and pretreated with Tris/EDTA/Borat Buffer pH 9.0 for 30 minutes at 100 °C.
Automatic staining was performed on a Leica Bond RX platform. Mouse monoclonal antibody clone 1E6 against the HEV ORF2 protein was incubated for 1 h at a dilution of 1:125 followed by a mix of Alexa Fluor 546-conjugated goat anti-mouse antibody (Invitrogen BV, A11018) and FITC-conjugated Rabbit anti-Human IgG (Gamma chain, Diagnostic Biosystem, F008) for 1 h at a dilution of 1:50. Following automated staining, the slides were hand washed in distilled H2O. Tissue was covered with Vectashield® Antifade Mounting Medium with DAPI (Vector Laboratories, H-1200), covered with a coverslip and stored at 4 °C until evaluation.
For single immunofluorescence on fresh frozen sections, standard procedures at the Department of Pathology and Molecular Pathology were applied using the Leica Bond automated staining system with IgG, IgA, IgM, kappa and lambda, C3, C4d antibodies (Diagnostic Biosystem, F008, F007, F009, F001 and F002, F003, BI-RC4D respectively, dilution: 1:25 for all antibodies).
Transmission electron microscopy (EM)
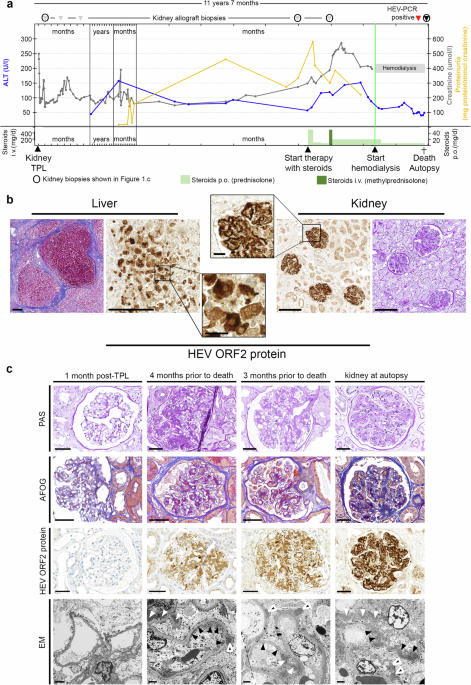
Archived images from electron microscopy performed on the allograft kidney biopsies of Patient 1, taken 11 years and 3 months (biopsy 3) as well as 11 years and 4 months post-transplantation (post-TPL) (biopsy 5) were re-evaluated. Additional ultra-thin sections from the archived grids were cut, stained and analyzed in a HITACHI TEM type H-7650 electron microscope.
Tissue samples of the allograft kidney from Patient 1 were collected at autopsy and were fixed in 2.5% buffered glutaraldehyde for at least 12 h, post-fixed for 2 h in 1% osmium tetroxide, dehydrated in a series of graded ethanol solutions and propylene oxide and embedded in epoxy resin 48 h at 60 °C. For comparison purposes, HEV negative tissue samples from the biopsy 1 (1 month post-TPL) of Patient 1 were sourced from archive FFPE material.
Semi-thin sections (1 µm) were stained with methylene blue – azure. Ultra-thin sections (90 nm) were stained with UranyLess and lead citrate. Sections were examined in a HITACHI TEM type H-7650 electron microscope.
In situ hybridization (ISH) for HEV RNA
In situ hybridization for HEV RNA was performed as described in ref. 40 using a commercially available probe designated V-HEV, targeting nucleotides 19-7257 of HEV-1 to -4 (Advanced Cell Diagnostics, #468111), a human negative (Dap-B) and a positive control (PPIB) –probes (Advanced Cell Diagnostics, #310043 and #313901 respectively). PPIB gene is expressed at low level in most of the tissues, between 10 to 30 copies per cell, providing a strict control for sample quality and technical performance.
Laser capture microscopy (LCM)
To perform mass spectrometry analysis of glomerular extracts from Patient 1, glomeruli stained for HEV ORF2 protein (1E6 antibody) were dissected using the ArcturusXT™ LCM System (Thermo Scientific). Protein extracts from one non-HEV-infected human kidney were used as control.
Serial cuts of the formalin-fixed paraffin-embedded kidney specimen from Patient 1 were prepared for LCM: one 2 µm thick section stained using 1E6 antibody, two 10 µm thick sections stained using Cresyl violet placed on polyethylene naphthalate (PEN) membrane glass slide and kept overnight in 4 °C44,45. Areas of interest were identified by overlapping the HEV ORF2+ glomeruli with the matching structures on the 10 µm thick section on PEN membrane glass slides. Two 10 µm thick sections from the non-HEV infected human kidney patient were similarly processed for glomeruli isolation. Collection of glomeruli was verified by microscopic examination of the LCM cap as well as the excised region (Fig. 3c). Two caps containing 40-50 glomeruli each were collected per case (one cap per 10 µm thick section, 2 sections per sample type, 4 caps in total). One cap containing interstitial tissue was also prepared per sample type (kidney from Patient 1 and the non-HEV infected patient, 2 caps in total). After excision, the caps containing tissue were transferred to a 1.5 mL centrifuge tube (Eppendorf ®Safe-Lock tubes) and frozen at −20 °C until further processing. As additional controls, liver samples from Patient 1 and non-HEV infected patient were similarly prepared.
Laser-captured sample preparation for mass spectrometry-based protein identification
For protein extraction, sterile blades and forceps were used to peel off the thermoplastic membranes containing laser-captured glomeruli or interstitial tissue from the LCM caps, which were then transferred into a sterile Eppendorf® Safe-Lock tube.
Eight samples in total (n = 4 for laser-captured glomeruli, n = 2 for laser-captured interstitial tissue, n = 2 for laser-captured liver tissue, from Patient 1 and non-HEV infected patient) were analysed at the Functional Genomic Center Zurich for mass spectrometry (MS) analysis. Briefly, the samples were lysed by adding 4% SDS lysis buffer and incubated for 60 minutes at 95 °C, 1 minute HIFU (High Intensity Focused Ultrasound), 10 minutes sonication, 30 minutes of additional heating at 95 °C, HIFU, sonication and max spin for 5 minutes, and then digested using trypsin solution on a thermo shaker at 37 °C overnight. The digested samples were dried and dissolved in 3% aqueous acetonitrile and 0.1% formic acid before being transferred in vials for liquid chromatography-mass spectrometry analysis. For the kidney samples, 200 ng peptides were injected on a M-class UPLC coupled to an Exploris Orbitrap mass spectrometer (ThermoFisher). The liver samples were loaded on evotips (Evosep) following the provided instructions and analyzed on an Evosep One LC coupled to a TIMS TOF Pro mass spectrometer (Bruker). The acquisition was done in diaPASEF mode.
The acquisition method used contained an inclusion list for the protein of interest, HEV ORF2 protein. For the kidney samples, the acquired MS data were processed for identification using the Maxquant search engine (V2.0.1.0). The spectra were searched against the HEV ORF2 protein sequence merged with the Homo sapiens protein database Swissprot, and also against interstitial markers (cytokeratin 7, complement 3, and collagen 1A1) and specific glomerular marker podocin46,47. For the liver samples, the acquired data independent acquisition spectra were processed with DIA-NN (Version 1648,) using a library free approach with the same protein database as for Maxquant. Results were summarized in the Proteome Software Scaffold (V 5.0.1) under very stringent settings (1% protein false discovery rate (FDR), a minimum of 2 peptides per protein, 0.1% peptide FDR) and expressed as averaged total spectrum counts ± s.d. (Supplementary Table 2).
Image processing
Slides with histologic / histochemical stains, immunohistochemistry and in situ hybridization were evaluated by light microscopy and scanned with a resolution of 0.250 µm per pixel (on a Hamamatsu Nanozoomer 2.0 HT whole slide imager) for evaluation by digital microscopy (NDP.view 2 viewer software, Hamamatsu Photonics K.K., V2.7.52). Regions of interest were exported as tif. files from scanned slides, from images acquired from a Zeiss Axio Scope.A1 equipped with a Zeiss Axiocam 100 color camera controlled via ZEN 2.3 lite Software, or from images acquired with an Olympus BX43 microscope equipped with an Olympus DP-43 digital camera controlled via Olympus cellSens software.
Immunofluorescence images were acquired with an upright fluorescence microscope (AxioImager.Z2 controlled by ZEN Blue software; 89 North Photofluor LM-75 light source, and Axiocam 503 mono camera; Zeiss, Jena, Germany), equipped with the following objectives: 20x (NA 0.5, Plan-NEOFLUAR), 40x (NA 1.4 oil, Plan-APOCHROMAT), and 100x (NA 1.45 oil, Plan-APOCHROMAT) objectives. This setup provides an excellent spatial resolution (nominally about 200 nm lateral resolution in our study; pixel size was 45.4 nm for 100x objective) comparable to confocal microscopy49. High resolution images were taken with the 100x objective using the ApoTome.2 module with deconvolution (grid 5 lp/mm; section thickness 0.7 µm). We used Vysis Abbott Chroma filter sets (Blue: excitation (ex) 335–383 nm, emission (em) 420–470; green: ex 481–507 nm; em 521–551 nm; red: ex 534–556 nm, em 574–606 nm). Control experiments with separate single-channel IgG or HEV ORF2 staining revealed negligible cross-talk between green and red fluorescence channels.
Co-localization of IgG and HEV ORF2 was quantified using Fiji50 and the JACoP ImageJ plug-in51. For analysis of entire glomeruli, images taken with the 20x objective were cropped to a rectangle around the glomeruli’s outlines. For the 100x images, subareas within individual glomeruli of 45-85 µm side lengths were selected. Pearson’s correlation coefficient (PCC) was calculated using Costes approach for automatic thresholding52,53. To test for significance of PCCs we used Costes randomization (1000 rounds) on each image pair with block sizes of 4 pixel (0.908 µm) for 20x and 10 pixel (0.454 µm) for 100x. P-values were all highly significant (p < 10E-10). We also calculated the Manders coefficients M1 and M254, which for Patient 1 were between 0.64 and 0.98 for 20x images (M1 = 0.84 ± 0.08, M2 = 0.83 ± 0.09; mean ± s.d., n = 25 glomeruli) and between 0.22 and 0.83 for 100x images (M1 = 0.58 ± 0.14, M2 = 0.72 ± 0.12; mean ± s.d.), consistent with high degrees of co-localization. Thresholds for M1 and M2 analysis were set manually.
For ultrastructural evaluation, ultrathin sections were examined using a transmission electron microscope (HITACHI H-7650). Regions of interest were exported in tif format. Figures were created using Adobe Photoshop software.
Western blotting
Autopsy kidney transplant and liver tissue samples from Patient 1 and normal kidney tissue sample from non-HEV-infected patient were homogenized in RIPA buffer (Sigma-Aldrich, R0278-50ML) containing phosphatase inhibitor and protease inhibitor cocktail tablets. Protein extracts were separated by SDS-PAGE on a 4-20% precast polyacrylamide gel (BioRad, #4561094) transferred to a PVDF membrane according to the manufacturer’s protocol and analyzed by Western blotting. The antibodies used were directed against HEV ORF2 protein (Millipore Corporation, MAB8002), PAX8 (Proteintech,10336-1-AP), Arginase-1 (Cell Marque, SP156), and β-Actin (Cell Signaling, 4970 T/S). Protein expression was detected by chemiluminescence and gel-imaging system (Bio Rad).
For PNGase F treatment, samples were denaturated for 10 minutes at 95 °C in glycoprotein denaturing buffer (New England Biolabs, NEB). Digestion with Peptide-N-Glycosidase F (PNGaseF, NEB, P0709,) was carried out for 4 h at 37 °C in the presence of 1% NP40 and the buffer provided by the manufacturer (NEB). Samples prepared without glycosidase but otherwise under the same conditions were used as controls.
Uncropped blots are provided in the Source Data file.
Molecular testing: qRT-PCR for HEV
For HEV RNA detection, a qRT-PCR protocol was applied to fresh frozen and FFPE tissue specimens. RNA was extracted with the Maxwell RSC simply RNA Kit (for frozen samples) and the Maxwell RSC RNA FFPE kit (for FFPE samples)(Promega) on a Maxwell instrument (Promega) according to the manufacturer’s recommendations.
cDNA reverse transcription was carried out using High Capacity cDNA Reverse Transcription Kit (Thermo Fischer) and following manufacturer’s recommendations. Briefly, reaction was done in 20 µL using random hexamer and 100 ng of total RNA as template. The cDNA samples were diluted twice for subsequent real-time PCR.
Real-time PCR has been performed as described in Fraga et al.55 using 2x TaqMan® Universal Master Mix II no UNG (Thermo Fischer) and a Quantstudio 5 apparatus (Thermo Fischer). Each 20-µL PCR reaction was performed with 5 µL of diluted cDNA. A standard curve prepared with dilutions of pUC-HEV-83-2-27 plasmid56 served to determine HEV genome copy number detected in each tissue sample.
A sample was considered as HEV positive if the average Ct value of the HEV amplicon was below the average Ct value of 35, corresponding to 102 copy/mL.
Molecular testing: HEV genotyping
Plasma sample was subjected to HEV genotyping following amplification of HEV ORF2 (592 bp) sequence, as described55. Genotype and subtype assignment were performed using the HEVnet genotyping tool (https://www.rivm.nl/mpf/typingtool/hev/).
Serological assay for anti-HEV IgG and IgM
Serum sample obtained in Cantonal Hospital Aarau was subjected to VIDAS® ANTI-HEV IgG and IgM assay (BioMérieux, France). Sample from University Zürich Hospital was assessed using Enzyme ImmunoAssays for the determination of IgG and IgM antibodies to hepatitis E Virus provided by Dia. Pro Diagnostic Bioprobes Srl (Italy).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.